Building Blocks for AI: Unpacking the Essentials of Test Data for Computer Vision in Digital Pathology (P.2) — Summary from An Excellent Review
Once we trained our computer vision (CV) models on an adequate and well-qualified training sample [1] of Whole Slide Images (WSIs), it is important to be fair in judging the model with “good” test data.
In this mini-review, I also distill the key characteristics of well-qualified test data, summarizing from Homeyer’s narrative review [2]. This excellent review provides us with recommendations for “good” test data.
 Overview of recommendations on compiling test datasets. Image from the paper [2].
Overview of recommendations on compiling test datasets. Image from the paper [2].
Recommendations
Homeyer’s review provided useful recommendations on many issues when creating a test dataset, including:
Target Population of Images. Before testing the model, we collect the data so that it represents the population of the model’s intended use. It means the data should be large enough to cover (at least) major types of variability. Different histologic patterns in a disease comprise biological variability while a variation in protocols of tissue processing leads to technical variability. Disagreement in evaluation among pathologists is the main source of observer variability.
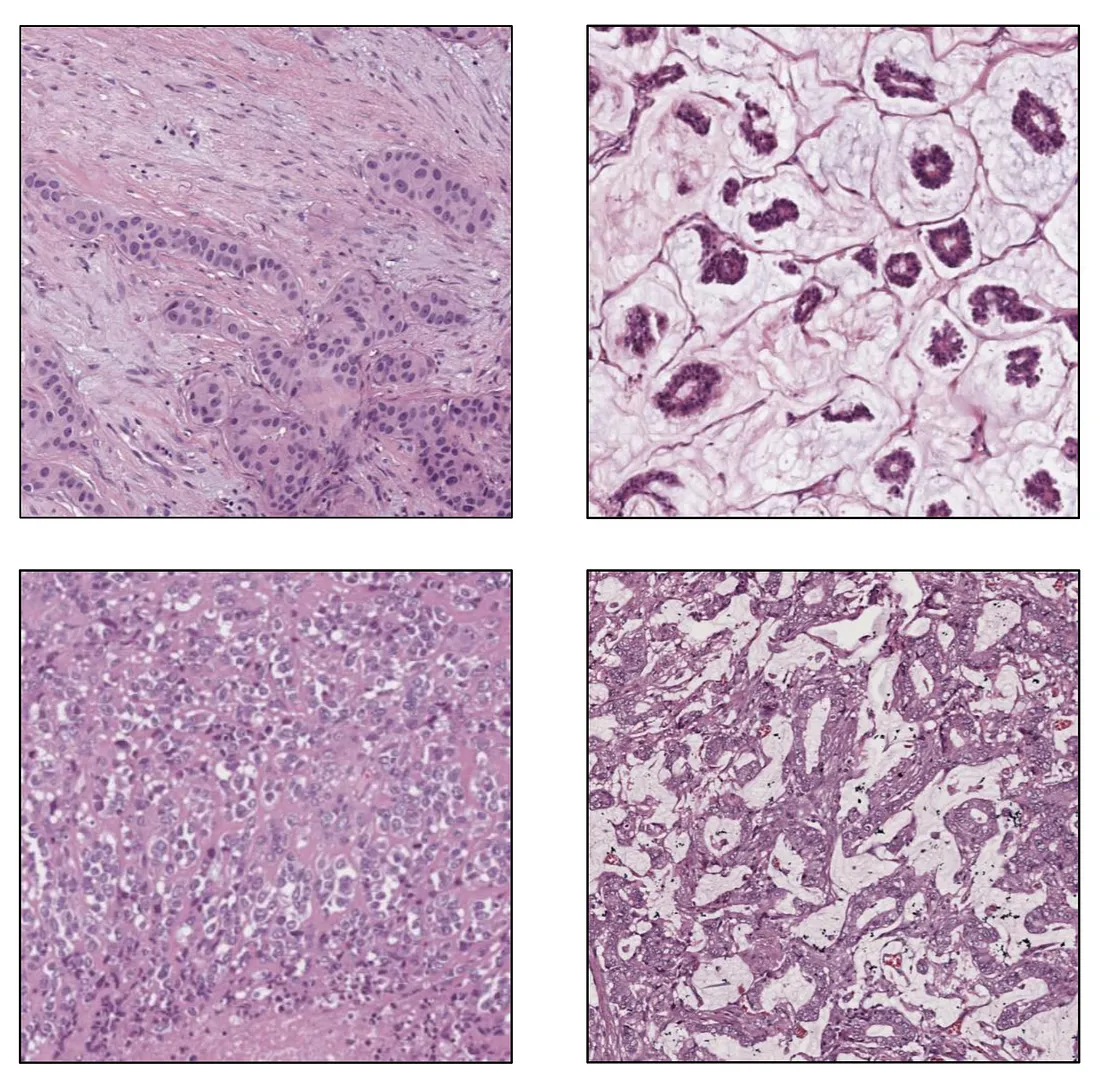 Biological Variability. Image from the author.
Biological Variability. Image from the author.
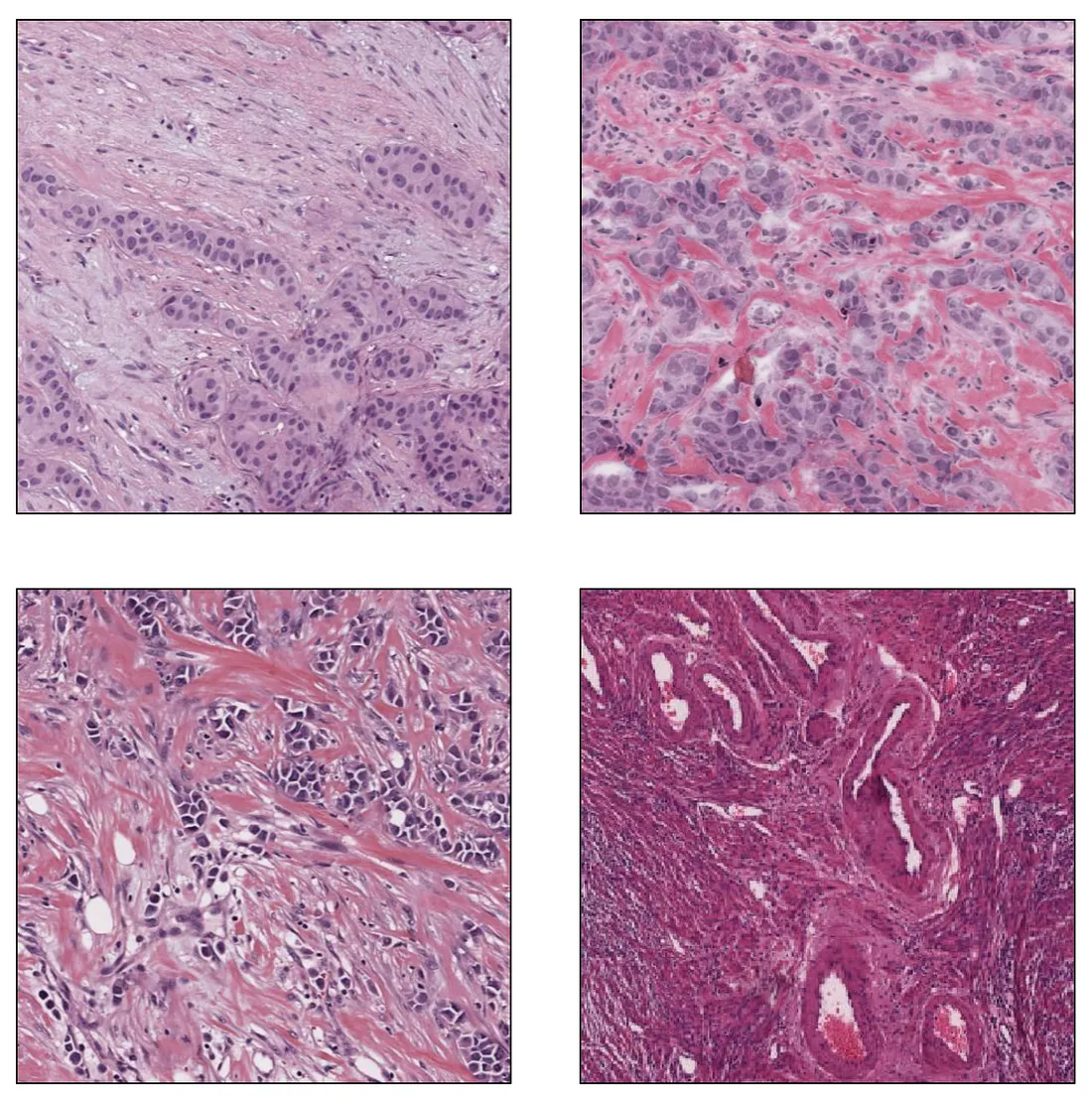 Inter-institutional staining variability, a type of Technical Variability. Image from the author
Inter-institutional staining variability, a type of Technical Variability. Image from the author
Data Collection. To create representative test data, we include all images encountered in the normal laboratory workflow. The general way is to collect all samples in a long enough period (enough cases are accumulated) and from multiple institutions.
Sample Size. Sample size directly affects sampling error. A larger sample size results in a smaller sampling error. We would want to consider the minimum sample size with an acceptable sampling error. “How much is an acceptable sampling error?” depends on the research question.
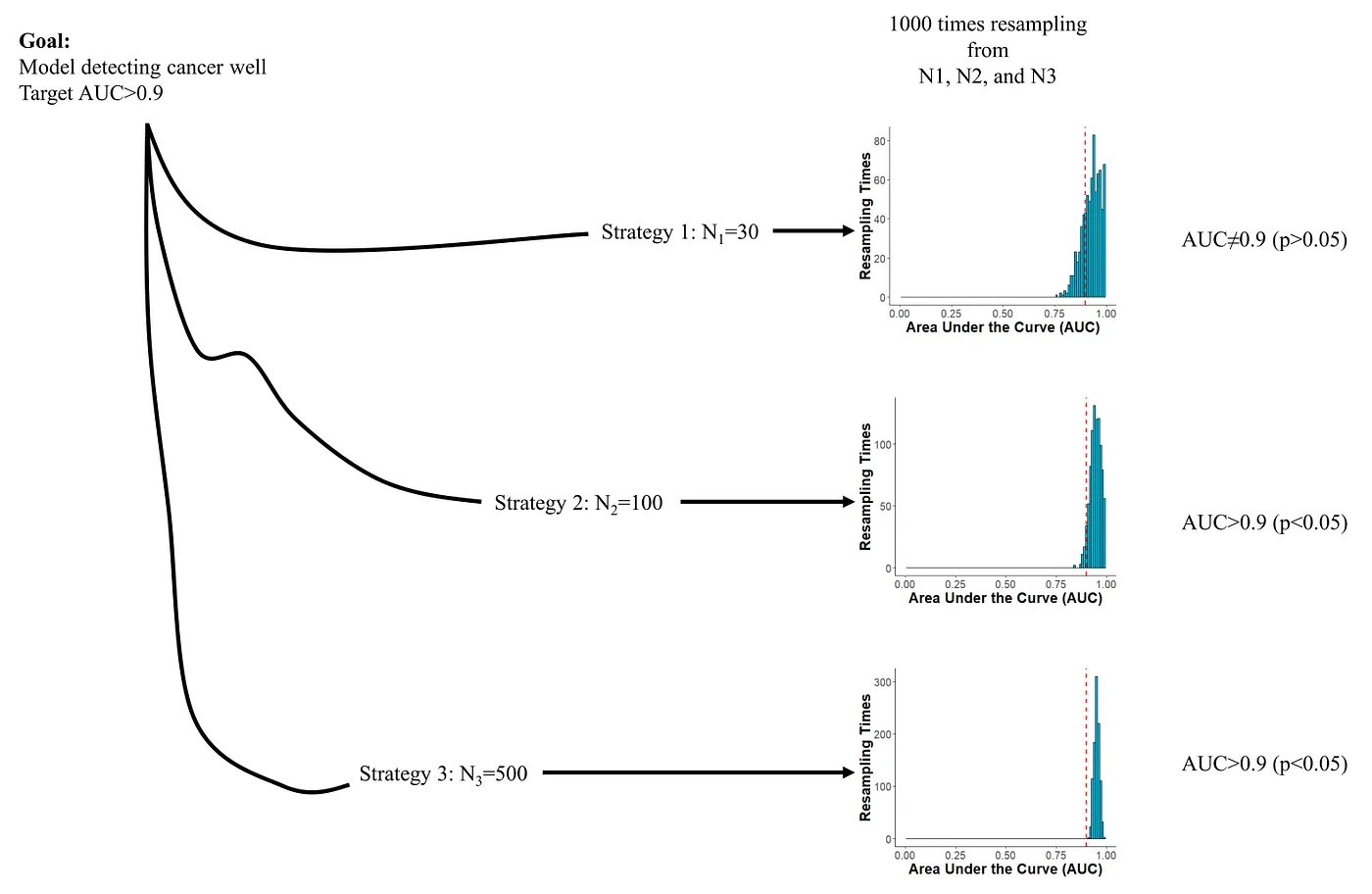 Accuracy and sample size trade-off. The confidence interval of AUC includes 0.9 in N1 and is higher than 0.9 in N2 and N3 but N3 is much more expensive to obtain. In a word, N1 is too few and N3 is too many. Image created by the author.
Accuracy and sample size trade-off. The confidence interval of AUC includes 0.9 in N1 and is higher than 0.9 in N2 and N3 but N3 is much more expensive to obtain. In a word, N1 is too few and N3 is too many. Image created by the author.
Subset. Some subsets of the data downgrade the model’s performance. This phenomenon is called hidden stratification. To detect hidden stratification, we can do subpopulation analysis (by known features such as clinical stages) and unsupervised clustering. The presence of hidden stratification requires further evaluation.
 Subsets can lead to hidden stratification. Image from the paper [2].
Subsets can lead to hidden stratification. Image from the paper [2].
Bias Detection. The paper refers to sampling bias. Sampling bias occurs when we do not randomly pick the samples by chance. Like we have types A and B in the population with a ratio of 1:1, we unfortunately pick 10 samples with 9 type-A and 1 type-B (which should be 5:5). For structured data, examining distributions of features can be helpful. For unstructured data (images), we can evaluate the distribution of the complex distributions extracted by cross-domain pretrained CV models.
Independence. It means that train and test datasets should not share a similar “spurious correlation”. Consider a scenario where we have n=100 samples from a single institution. We randomly split them into train (n=80) and test (n=20) sets. Despite the fact that the test set comprises unique samples distinct from the train set, they originate from the same institution and are therefore subject to the same “spurious correlations”. Therefore, good performance in this “internal” test set could be just an illusion. Testing the model with an independent, “external” test set (like in another institution) is always a good idea.
Reporting. An adequate report of test data features is crucial for various purposes. Some important points to report are:
- Metadata
- Data acquisition protocol
- Annotation protocol (including software and hardware)
- Summary statistics
These reports are important to detect bias and subset to qualify the test data.
Regulatory Requirements. Regulatory approval by the U.S. Food and Drug Administration (FDA) and European Union (EU) is required for sale and clinical use.
References
- Le, Minh-Khang. “Building Blocks for AI: Unpacking the Essentials of Training Data for Computer Vision in Digital Pathology (P.1)” Medium, 9 Dec. 2023, medium.com/p/f5b9e8ec7b39.
- Homeyer, André, et al. “Recommendations on Compiling Test Datasets for Evaluating Artificial Intelligence Solutions in Pathology.” Modern Pathology, vol. 35, no. 12, 1 Dec. 2022, pp. 1759–1769.