Embarking on a Journey: A Novice’s Guide to Histopathology Annotation in Deep Learning
Indigital pathology research, annotation serves an important role in the pipeline. It can bring failures to the research if ignored. The way to annotate depends on the context and the task at hand. In this mini-review, I share my experience with types of histopathology annotation in digital pathology research.
Categories of annotation
Annotation for Segmentation. The process of annotation for segmentation is a meticulous task, which consumes a lot of human effort, especially when performed manually. Ideally, the pathologists have to annotate pixel to pixel in order to train the model properly. However, given the vast dimensions of whole slide images (WSIs) and the demanding schedules of pathologists, this approach is practically unfeasible. Therefore, alternative ways are required. Nuclei segmentation [1] necessitates only point annotation, from which the algorithm generates pixel-level annotation.
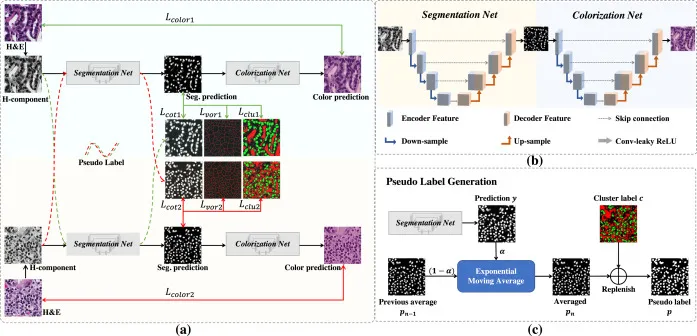 Nuclear Segmentation pipeline (Image from the original paper 10.1016/j.media.2023.102933)
Nuclear Segmentation pipeline (Image from the original paper 10.1016/j.media.2023.102933)
Using a distinct approach, the SegPath [2] pipeline uses immunofluorescence staining on de-stained HE slides to make labels.
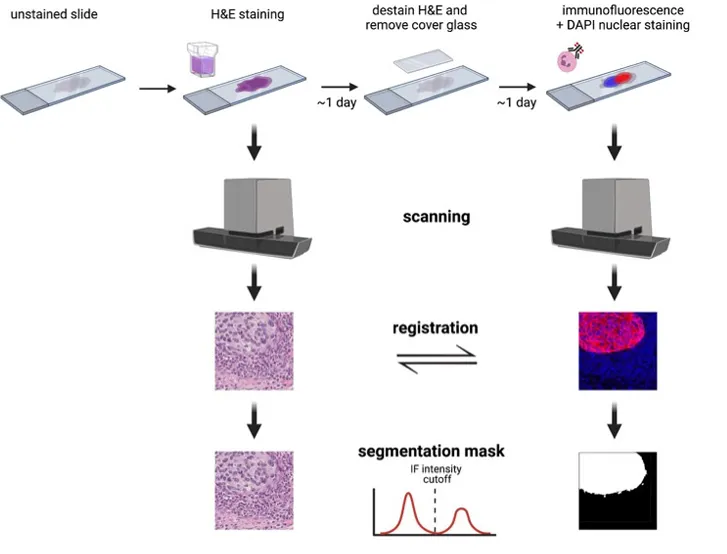 SegPath pipeline (Image from the original paper: 10.1101/2022.05.09.489968)
SegPath pipeline (Image from the original paper: 10.1101/2022.05.09.489968)
Detailed explanations of these approaches are out of our scope.
Annotation for Other Tasks. This annotation depends on how we design the task, which includes classification, regression, and survival prediction. When our objective is for the model to focus on the tumor region, we annotate precisely that — the tumor region, deliberately excluding the space and normal tissue. Similarly, if our goal is to highlight high-grade locations, we annotate these specific areas, consciously excluding space, normal tissue, and low-grade tumor regions. This method guides the model to concentrate on the high-grade areas, providing a nuanced understanding of the task at hand.
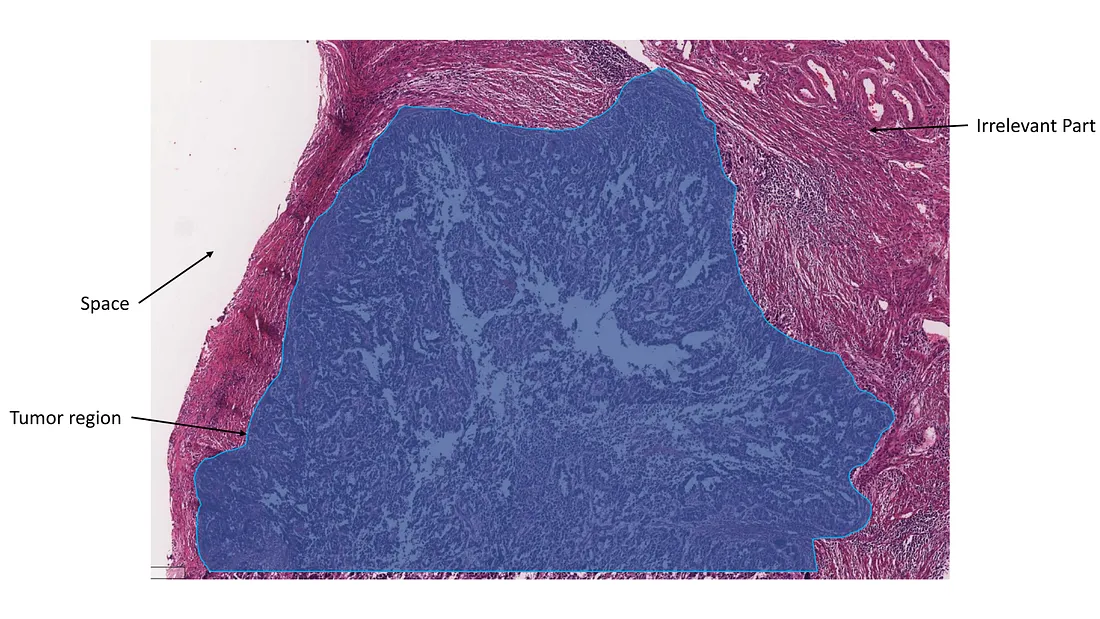 Tumor region annotation, excluding space and irrelevant tissue (Image created by the author)
Tumor region annotation, excluding space and irrelevant tissue (Image created by the author)
Annotation for Quality Control. This annotation is designed to enhance the quality control process by minimizing artifacts. It is a relatively feasible approach in terms of human effort, yet it can significantly improve the quality of training and validation data. This, in turn, leads to enhanced model performance and reliability.
No Annotation. No-annotation algorithms have also been developed. This enables the model to interact with the WSI without the need for annotation, reducing human effort. The downside is that it is susceptible to artifacts and pitfalls, which can increase the “black box” features of the model. Therefore, the model often incorporates automatic algorithms to eliminate irrelevant or misleading parts of the WSI. Techniques such as attention mechanism, feature selection, or clustering are employed for this purpose.
Conclusion
Annotation is an important part of the deep-learning pipeline in digital pathology. The annotation strategy depends on the design of the model and directly influences the quality of training. In essence, it’s a process that guides the model to focus on relevant features, akin to a scientist highlighting significant data points in an experiment.
To those who may be interested, I hope you enjoy this mini-review.
Reference
- Lin, Yi, et al. “Nuclei Segmentation with Point Annotations from Pathology Images via Self-Supervised Learning and Co-Training.” Medical Image Analysis, vol. 89, 1 Oct. 2023, p. 102933, doi https://doi.org/10.1016/j.media.2023.102933.
- Komura D, et al. “Beyond pathologist-level annotation of large-scale cancer histology for semantic segmentation using immunofluorescence restaining”. bioRxiv. doi: 10.1101/2022.05.09.489968.