Label Noise and Its Problems for Computer Vision Model in Histopathology
Introduction
Label noise refers to inaccuracies or errors in the labels of a dataset in the context of deep learning.
In this mini-review, I would like to point out the inevitable presence of label noise in histopathology annotation and labeling. The awareness of pathology label noise can significantly improve the quality of the training process and the model itself.
In this mini-review, we will address 4 bullet questions:
-
What are the types of label noise?
-
What are the main sources of label noise in the pathology context?
-
What are the effects caused by label noise?
4.What are the main solutions to the label noise problem?
Types of Label Noise
To better understand the types of label noise, we considered the following example: a pathologist is asked to label 100 histopathological patches into 4 divided classes, including tumor glands, normal glands, tumor-related stroma, and normal stroma.
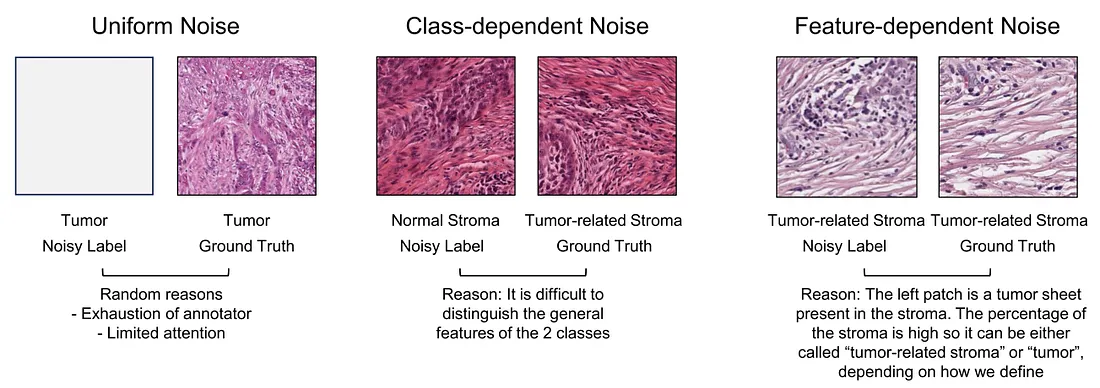 The label noise is divided into 3 main categories [1], as follows:
The label noise is divided into 3 main categories [1], as follows:
Uniform noise. This type of noise is not related to any class and it happens completely random. In our example, if the pathologist is tired from practical work, he/she mislabels some patches randomly.
Class-dependent noise. This noise happens due to the general difficulty of distinguishing the classes. In the above example, the pathologist is confused to distinguish the tumor-related stroma and normal stroma. It is because they are basically similar in histopathology if there is no sign of tumor cells in the patch. “It is generally difficult” is a sign of class-dependent noise.
Feature-dependent noise. This type occurs when the probability of a label being noisy depends on the features of the patch. In the above example, our pathologist finds that some glandular structures are intermediate between tumor and normal gland, which confuses him/her. He/she may mislabel some of them because of their confusing features. “It is difficult only in some cases” is a sign of feature-dependent noise.
These 3 types, by definition, differ from each other by the causes of increasing probability of the label being noisy. The distributions of these noise categories are also different. Moreover, there are noises that can belong to more than one category, which is much more complicated to handle.
Label Noise in Pathology
Computer vision in pathology usually requires the whole slide image (WSI) to be cropped into smaller patches because WSI is too large to be handled as a single input. Annotation (manual or automated) is usually applied to improve the efficiency of the cropping process because a significant portion of WSI contains uninformative features, such as spaces, hemorrhages, and so on. The patch-based approach, along with the annotation, potentially introduces label noises into the data.
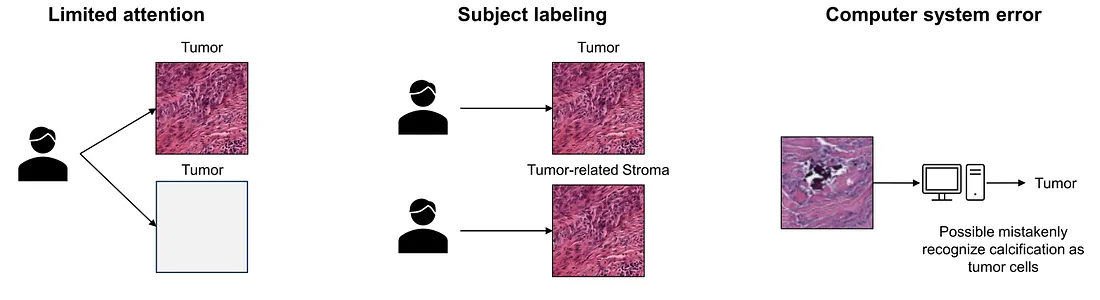
In this discussion, I will summarize the main causes of label noise in histopathology computer vision of which I am aware. In medical image data, label noise originates from [2]:
(1) The limited attention or expertise of the human annotator. This can be different types of noise. For example, if the limited attention simply comes from the exhaustion of the annotator, it is a uniform noise. On the other hand, if the 2 classes cannot be distinguished by the expertise of the annotator, meaning if the annotation is handed to an annotator with higher expertise and experience, these 2 classes can be annotated more correctly. Then, it is a class-dependent label.
(2) Subjective nature of labeling. This creates inter-observer variation. The consensus is easy to reach in some cases while labeling can be very challenging in other cases. This is considered as feature-dependent noise.
(3) Errors in computerized labeling systems. The source of label noise in this situation also depends on the underlying cause of the erroneous behavior of the computerized system.
Damage caused by label noise
Too much label noise causes the model to overfit easily, therefore, leading to model performance degradation. Label noise can direct the gradient and make it impossible to understand the results of model interpretability analysis.
Solutions to label noise
There are many approaches to tackle the label noise problem. The obvious way is to clean the data, but as we mentioned before, it is sometimes challenging to define the clean label by human annotators, even by experts, in feature-dependent label noise. Therefore, the problem is not that easy. Some approaches have been proposed:
-
Data cleaning [3]. This can be done by manual double-check or automatic correction algorithm.
-
Loss function modifications [4]. These losses are designed or modified (from known loss) to make it robust to noisy labels.
-
Model regularization [2]. The regularization techniques are basically used to improve the model’s robustness to various factors, including label noise. Drop-out, weight decay, and image augmentation are well-known examples.
-
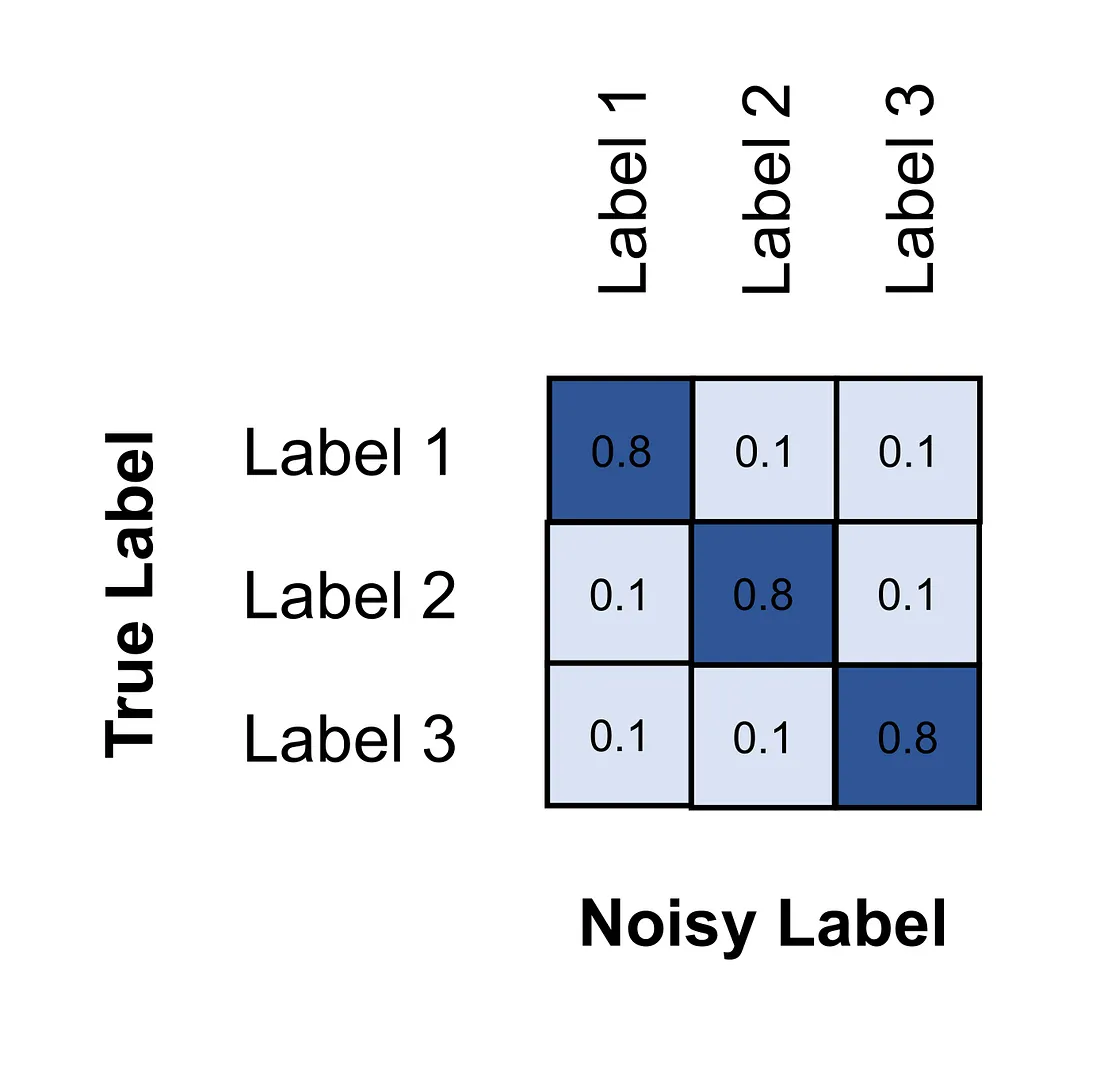
Noise modeling [3]. This technique requires us to add and investigate the noise transition matrix that captures the probability of each label flipped into another label.
- Semi-supervised learning [3]. This learning scheme involves a scenario where there are 2 datasets: small but clean data and large but noisy data. By utilizing these 2 datasets, one develops an approach to improve the model’s robustness to label noise.
Conclusion
In summary, label noise is a challenge in pathology computer vision projects because of its inevitable presence and damage to the model’s performance. We conclude our discussion by addressing the bullet questions:
- What are the types of label noise?
There are 3 main types of label noise, which are uniform noise, label-dependent noise, and feature-dependent noise.
- What are the main sources of label noise in the pathology context?
There are many sources of label noise, mainly originating from (1) the limited attention or expertise of human annotators, (2) the subjective nature of labeling, and (3) errors in computerized labeling systems.
- What are the effects caused by label noise?
Label noise causes overfitting, model performance degradation, and makes it difficult to interpret the model’s decision.
- What are the main solutions to the label noise problem?
There are 5 main solutions to the problem. Data cleaning is the most important but challenging solution. Others include modifying loss function, model regularization, noise modeling, and semi-supervised learning.
To those who may be interested, I hope you enjoy this mini-review.
Reference
-
Algan, Görkem, and İlkay Ulusoy. “Label Noise Types and Their Effects on Deep Learning.” ArXiv.org, 23 Mar. 2020, arxiv.org/abs/2003.10471. Accessed 4 Oct. 2023.
-
Karimi, Davood, et al. “Deep Learning with Noisy Labels: Exploring Techniques and Remedies in Medical Image Analysis.” Medical Image Analysis, June 2020, p. 101759, https://doi.org/10.1016/j.media.2020.101759.
-
Li, Shao-Yuan, et al. “Improving Deep Label Noise Learning with Dual Active Label Correction.” Machine Learning, vol. 111, no. 3, 6 Jan. 2022, pp. 1103–1124, https://doi.org/10.1007/s10994-021-06081-9. Accessed 11 Aug. 2022.
-
Kim, SeongKyung. “Handling Noisy Label Data with Deep Learning.” MLearning.ai, 26 Nov. 2022, medium.com/mlearning-ai/handling-noisy-label-data-with-deep-learning-ff986deedc76.